※ DOG INTRODUCTION:
Schematic diagram of protein domain structures with functional motifs/sites is an important part in a large proportion of scientific articles which are focused on molecular and cellular researches. In order to estimate how many papers will contain protein domain graphs, we simply chose five typical molecular and cellular magazines, including Cell, Nature Cell Biology (NCB), The Journal of Cell Biology (JCB), Molecular Biology of the Cell (MBC) and Molecular and Cellular Biology (MCB). Then we carefully read all research articles of the five journals published in 2007, while the reviews and other types of articles were not taken into account. Totally, there were 551 of 1994 articles (27.6%) to contain at least one figure for protein domain structures. Thus, diagram of protein domain/motif structures in an attractive, concise and precise manner is greatly helpful for a broad readership to grasp the old and novel functions of proteins rapidly.
In this work, we present a novel software of DOG (Domain Graph, version 1.0) for experimentalists, to prepare publication-quality figures of protein domain structures. The scale of a protein domain and the position of a functional motif/site will be precisely calculated. The DOG 1.0 was written in JAVA 1.5 (J2SE 5.0) and packed with Install4j 4.0.8. Thus, the DOG 1.0 could be easily installed on a computer. Then we developed several packages to support three major Operating Systems (OS), including Windows, Unix/Linux and Mac. The Windows XP, Fedora Core 6 OS (Linux), Apple Mac OS X 10.4 (Tiger) and 10.5 (Leopard) were chosen to test the stability of DOG1.0 software. For Windows and Linux systems, a Java Runtime Environment 6 (JRE) package of Sun Microsystems was also included. The DOG 1.0 software is freely available from: http://dog.biocuckoo.org.
DOG 1.0 User Interface
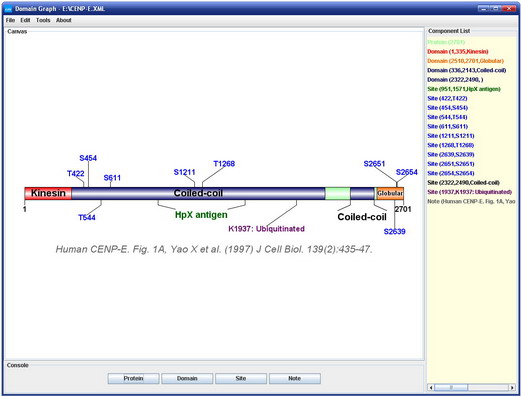
For publication of results please cite the following article:  DOG 1.0: Illustrator of Protein Domain Structures.
DOG 1.0: Illustrator of Protein Domain Structures.Jian Ren, Longping Wen, Xinjiao Gao, Changjiang Jin, Yu Xue and Xuebiao Yao. Cell Research (2009) 19:271–273. |



